Comparing Feature Selectors
Hi! You want to compare the performance of multiple feature selectors? This is an example Notebook, showing you how to do such an analysis.
Prerequisites
We are going to use more or less the same configuration as we did in the Quick start example, but then with more Feature Selectors. Again, start by downloading the example project: comparing-feature-selectors.zip
Installing the required packages
Now, let's install the required packages. Make sure you are in the comparing-feature-selectors folder, containing the requirements.txt file, and then run the following:
pip install -r requirements.txt
Running the experiment
Run the following command to start the experiment:
python benchmark.py --multirun ranker="glob(*)" +callbacks.to_sql.url="sqlite:////tmp/results.sqlite
Analyzing the results
There now should exist a .sqlite file at this path: /tmp/results.sqlite:
```
$ ls -al /tmp/results.sqlite
-rw-r--r-- 1 vscode vscode 20480 Sep 21 08:16 /tmp/results.sqlite
```
Is that the case? Then let's now analyze the results! 📈
We will install plotly-express, so we can make nice plots later.
%pip install plotly-express nbconvert --quiet
Next, let's find a place to store our results to. In this case, we choose to store it in a local SQLite database, located at /tmp/results.sqlite.
import os
con: str = "sqlite:////tmp/results.sqlite"
con
'sqlite:////tmp/results.sqlite'
Now, we can read the experiments table.
import pandas as pd
experiments: pd.DataFrame = pd.read_sql_table("experiments", con=con, index_col="id")
experiments
| dataset | dataset/n | dataset/p | dataset/task | dataset/group | dataset/domain | ranker | validator | local_dir | date_created | |
|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||
| 3lllxl48 | My synthetic dataset | 10000 | 20 | classification | None | None | ANOVA F-value | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:28:27.506838 |
| 1944ropg | My synthetic dataset | 10000 | 20 | classification | None | None | Boruta | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:28:31.230633 |
| 31gd56gf | My synthetic dataset | 10000 | 20 | classification | None | None | Chi-Squared | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:29:19.633012 |
| a8washm5 | My synthetic dataset | 10000 | 20 | classification | None | None | Decision Tree | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:29:23.459190 |
| 27i7uwg4 | My synthetic dataset | 10000 | 20 | classification | None | None | Infinite Selection | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:29:27.506974 |
| 3velt3b9 | My synthetic dataset | 10000 | 20 | classification | None | None | MultiSURF | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:29:31.758090 |
| 3fdrxlt6 | My synthetic dataset | 10000 | 20 | classification | None | None | Mutual Info | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:35:04.289361 |
| 14lecx0g | My synthetic dataset | 10000 | 20 | classification | None | None | ReliefF | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:35:08.614262 |
| 3sggjvu3 | My synthetic dataset | 10000 | 20 | classification | None | None | Stability Selection | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:35:59.121416 |
| dtt8bvo5 | My synthetic dataset | 10000 | 20 | classification | None | None | XGBoost | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:36:23.385401 |
Let's also read in the validation_scores.
validation_scores: pd.DataFrame = pd.read_sql_table("validation_scores", con=con, index_col="id")
validation_scores
| index | n_features_to_select | fit_time | score | bootstrap_state | |
|---|---|---|---|---|---|
| id | |||||
| 3lllxl48 | 0 | 1 | 0.004433 | 0.7955 | 1 |
| 3lllxl48 | 0 | 2 | 0.004227 | 0.7910 | 1 |
| 3lllxl48 | 0 | 3 | 0.005183 | 0.7950 | 1 |
| 3lllxl48 | 0 | 4 | 0.003865 | 0.7965 | 1 |
| 3lllxl48 | 0 | 5 | 0.002902 | 0.7950 | 1 |
| ... | ... | ... | ... | ... | ... |
| dtt8bvo5 | 0 | 16 | 0.000670 | 0.7805 | 1 |
| dtt8bvo5 | 0 | 17 | 0.000480 | 0.7725 | 1 |
| dtt8bvo5 | 0 | 18 | 0.003159 | 0.7760 | 1 |
| dtt8bvo5 | 0 | 19 | 0.000848 | 0.7650 | 1 |
| dtt8bvo5 | 0 | 20 | 0.000565 | 0.7590 | 1 |
160 rows × 5 columns
We can now merge them. Notice that we set as the index the experiment ID, so we can use pd.DataFrame.join to do this.
validation_scores_with_experiment_info = experiments.join(
validation_scores
)
validation_scores_with_experiment_info.head(1)
| dataset | dataset/n | dataset/p | dataset/task | dataset/group | dataset/domain | ranker | validator | local_dir | date_created | index | n_features_to_select | fit_time | score | bootstrap_state | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||
| 14lecx0g | My synthetic dataset | 10000 | 20 | classification | None | None | ReliefF | k-NN | /workspaces/fseval/examples/comparing-feature-... | 2022-10-22 14:35:08.614262 | NaN | NaN | NaN | NaN | NaN |
Cool! That will be all the information that we need. Let's first create an overview for all the rankers we benchmarked.
validation_scores_with_experiment_info \
.groupby("ranker") \
.mean(numeric_only=True) \
.sort_values("score", ascending=False)
| dataset/n | dataset/p | index | n_features_to_select | fit_time | score | bootstrap_state | |
|---|---|---|---|---|---|---|---|
| ranker | |||||||
| Infinite Selection | 10000.0 | 20.0 | 0.0 | 10.5 | 0.004600 | 0.818925 | 1.0 |
| XGBoost | 10000.0 | 20.0 | 0.0 | 10.5 | 0.002998 | 0.818575 | 1.0 |
| Decision Tree | 10000.0 | 20.0 | 0.0 | 10.5 | 0.002810 | 0.817675 | 1.0 |
| Stability Selection | 10000.0 | 20.0 | 0.0 | 10.5 | 0.002406 | 0.803325 | 1.0 |
| Chi-Squared | 10000.0 | 20.0 | 0.0 | 10.5 | 0.002548 | 0.795975 | 1.0 |
| ANOVA F-value | 10000.0 | 20.0 | 0.0 | 10.5 | 0.003745 | 0.789275 | 1.0 |
| Mutual Info | 10000.0 | 20.0 | 0.0 | 10.5 | 0.002314 | 0.786475 | 1.0 |
| Boruta | 10000.0 | 20.0 | 0.0 | 10.5 | 0.002366 | 0.518075 | 1.0 |
| MultiSURF | 10000.0 | 20.0 | NaN | NaN | NaN | NaN | NaN |
| ReliefF | 10000.0 | 20.0 | NaN | NaN | NaN | NaN | NaN |
Already, we notice that MultiSURF and ReliefF are missing. This is because the experiments failed. That can happen in a big benchmark! We will ignore this for now and continue with the other Feature Selectors.
👀 We can already observe, that the average classification accuracy is the highest for Infinite Selection. Although it would be premature to say it is the best, this is an indication that it did will for this dataset.
Let's plot the results per n_features_to_select. Note, that n_features_to_select means a validation step was run using a feature subset of size n_features_to_select.
import plotly.express as px
px.line(
validation_scores_with_experiment_info,
x="n_features_to_select",
y="score",
color="ranker"
)
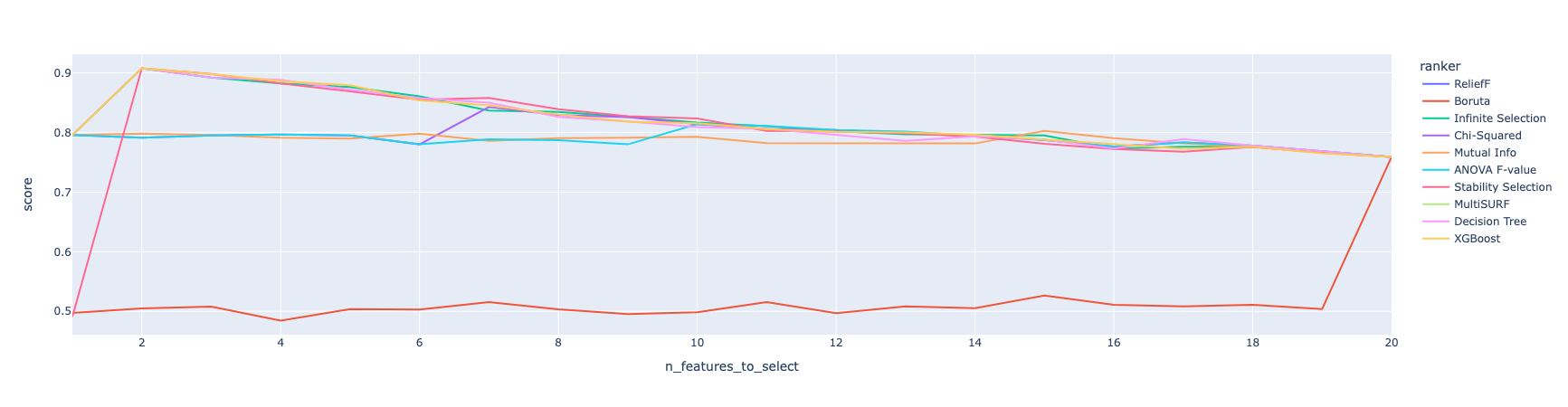
Indeed, we can see XGBoost, Infinite Selection and Decision Tree are solid contenders for this dataset.
🙌🏻
This has shown how easy it is to do a large benchmark with fseval. Cheers!