Getting started
To get started, there's two main resources.
The ⚡️ Quick start guide below 👇🏻
⚡️ Quick start
Let's run our first experiment. The goal will be to compare two feature selectors ANOVA F-Value and Mutual Info.
First, install fseval:
pip install fseval
If for some reason you would not like to install fseval from the PyPi package index using pip install like above, you can also install fseval right from its Git source. Execute the following:
git clone https://github.com/dunnkers/fseval.git
cd fseval
pip install -r requirements.txt
pip install .
You should now be able to continue in the same way as before ✓.
Now you can decide whether you want to define your configuration in YAML or in Python. Choose whatever you find most convenient.
- YAML
- Structured Config
Download the example configuration: quick-start-yaml.zip
Then, cd into the example directory. You should now have the following files:
Download the example configuration: quick-start-structured-configs.zip
You should now have the following files:
We can now decide how to export the results. We can upload our results to a live SQL database. For now, let's choose a local database. SQLite is good for this.
sql_con=sqlite:////Users/dunnkers/Downloads/results.sqlite # any well-defined database URL
If you define a relative database URL, like sql_con=sqlite:///./results.sqlite, the results will be saved right where Hydra stores its individual run files. In other words, multiple .sqlite files are stored in the ./multirun subfolders.
To prevent this, and store all results in 1 .sqlite file, use an absolute path, like above. But preferably, you are using a proper running database - see the recipes for more instructions on this.
We are now ready to run an experiment. In a terminal, cd into the unzipped example directory and run the following:
python benchmark.py --multirun ranker='glob(*)' +callbacks.to_sql.url=$sql_con
And our experiment starts running 👏🏻!
Using --multirun combined with ranker='glob(*)' we instructed fseval to run experiments for all available rankers. The results are now stored in a SQLite database.
$ tree ~/Downloads
/Users/dunnkers/Downloads
└── results.sqlite
0 directories, 1 file
We can open the data using DB Browser for SQLite. We can access the validation scores in the validation_scores table:
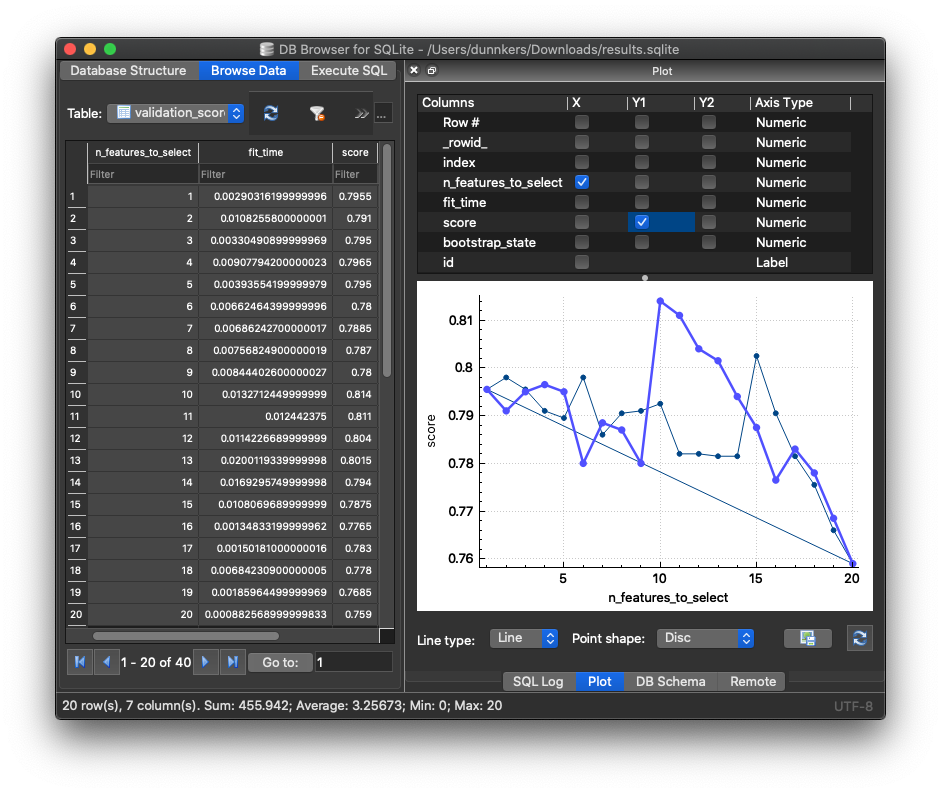
In the example above,
the graph plots the feature subset size (n_features_to_select) vs. classification accuracy (score).
For our two feature selectors, ANOVA F value vs. Mutual Info, we can now see which gets the highest classification accuracy with which feature subset. Using fseval, we can easily compare many feature- selectors or rankers, and at a large scale 🙏🏻.